In 1990, the Human Genome Project was launched as one of the great undertakings of scientific exploration with the objective to determine the DNA sequence of the entire human genome. After 13 years of collaboration and hard work by students and scientists around the world, by 2003 they finally had a semi-complete human genome. This achievement, which was at first received with a high degree of skepticism by scientists and the public alike, ushered in a new era of healthcare that continues to be helpful in developing novel cures for some of the most important diseases.
Advances in healthcare are partly due to the improvement in DNA sequencing technology and computational tools that allow the reconstruction of chromosome level assembly from fragmented DNA sequences. The exponential advancement of ‘computer power’ (as described by Moore’s law) greatly contributed to the low sequencing costs for the genome, allowing various companies such as DeepGenomics, 23&Me and Ancesry.com to solve challenging healthcare problems by deciphering the complex language of DNA.
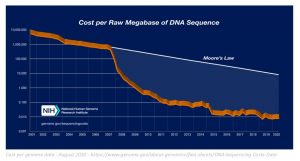
The decrease in costs for generating genomic data has given rise to data-driven fields such as precision medicine, genomic selection and gene editing. The field of gene editing, for example, relies on knowing where to edit the genome in order to obtain the desired phenotypes. This knowledge comes from increased accessibility of DNA. Although generating data costs were significantly reduced, managing this data and generating high quality information and insights from it, is still a bottleneck due to its computational complexity.
February 15, 2021, marked the 20th anniversary of the Human Genome Project. For plant crops, various consortiums were established, similar to the Human Genome Project, to elucidate the genetic architecture of plant species in order to devise better strategies for improved crop yield and quality. Among many, some of the most important collaborations were the Wheat Genome Project and the Maize Genome Project. Both projects laid the groundwork for significant improvements of these economically important crops.
The overarching goal for plant agriculture has always been to improve crop quality. This has been largely achieved by investigating genomic and phenotypic variations in crops of the same species. Traditionally, crosses of different phenotypes were made in order to assess the variation in the progeny and thus enable improved crop quality. A single representative reference genome for the organisms has generally worked well for species that are highly identical. However, this is rarely the case in plants. For example, there is great genomic diversity within the maize species where one-third of its gene content is not shared between varieties. In addition, the genomic divergence between two maize varieties is roughly equal to the divergence between humans and chimpanzees.
In cases such as maize, a single representative reference genome poses a challenge as it is unable to capture the full diversity of this unique organism. The solution to this is a pan-genome, which is an entire set of genes that exists in all members of the species. When studying plants, the differences in the genes, their presence or absence, gene variation or even the number of copies that they have, could contribute to various quality traits such as yield, disease resistance, and fruit production that would otherwise not be recognized with a single reference genome.
At NRGene, on the daily basis we dig into DNA data to reveal innovative insights that advance genomic knowledge, as such, we are happy to join in celebrating DNA day. NRGene has been involved in plant genomics for many years and, through our computational tools, we gained the capabilities to decipher plant DNA and use that data to bring plant science and variety improvement to a much higher level.
Happy DNA Day!