PanMAGIC™ builds a foundation for better breeding through whole genome analysis. NRGene’s PanMAGIC™ delivers mapping and gene space comparison between several assembled genomes of the same organism.
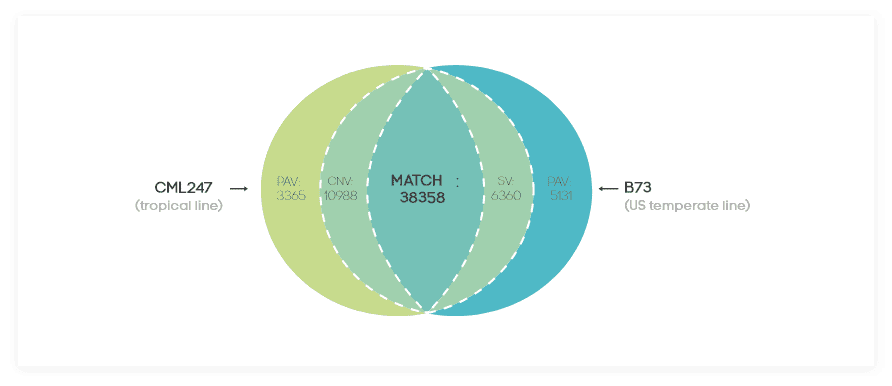
One reference genome is not enough to uncover complete diversity in species. For instance, independent studies comparing two maize variants have shown that half of the genome can be structurally different between variants. Empowered breeders need high volume and diverse data for comparison.
The whole genome analysis of PanMAGIC™ linearly maps full genomes, giving you complete genomic context and coordinate conversion. Unlike other methods, PanMAGIC™ includes all-to-all mapping. PanMAGIC™ reveals full genomic context allows robust exploration of genetic variations and eliminates bias towards one reference
Comparative genomics solution for whole genome analysis
PanMAGIC™ is a comparative genomic solution, making it an important part of the NRGene solutions. Pan-genome comparison is performed between several de novo assemblies of the same species. Instead of comparing a genome to one reference genome or specific loci, PanMAGIC™, performs a full genome-to-genome correlation. At NRGene, we are constantly enriching our database. We have established plant consortiums for key crops, including tomato, potato, wheat, ryegrass, and canola. The power of having multiple genomes for each crop, and the ability to compare them, translates into groundbreaking results that put NRGene at the forefront of plant genomics. With PanMAGIC™, researchers and breeders will be better equipped to identify genes for improving their crop production.
NRGene’s PanMAGIC™ takes full genome analysis to the next level, leveraging NRGene’s extensive and advanced algorithmics and proprietary database. PanMAGIC™ service performs a detailed comparative analysis to understand diversity, genomic changes, or structural variations between genomes. This level of detail has accelerated research, allowing researchers to describe a high level of diversity in their genetic lines. We use multiple assemblies to generate an understanding of the genome-to-genome correlation, as well as the position and visualization of multiple genomes. PanMAGIC is an important basis for better breeding, eliminating bias toward any reference variety and creating a robust infrastructure for cost-effective genomics and diversity analysis. From sweet potatoes, to commercial food potatoes, to cannabis, PanMAGIC™ can shed light on any specie.
Mapping and comparing several whole genomes to capture the full and true diversity within the species.
First, an all-to-all mapping of all the genomes is performed, generating multiple text files that include the mapping of each individual genome to every other genome. Further, transcriptomic data is mapped to each of the genomes. Combining the genomes and the transcriptome mapping, a pair-wise comparison of the genic content of the genomes is conducted resulting in detection of SV (structural variations, e.g. translocations), PAVs (Presence/Absence Variation) and CNVs (Copy Number Variations).

Outstanding Results