TraitMAGIC™ is NRGene’s solution for mapping QTLs meaning mapping phenotypes to genomic locations and specific genomic markers.
Reliable QTL mappings allows for faster and more cost-effective breeding processes.
By using TraitMAGIC, breeding companies will be able to map important traits such as the taste, aroma, and color in a range of organisms from maize and wheat to watermelons and trees.
Researchers value TraitMAGIC™ as a robust and validated tool for identifying unique markers relevant for multiple traits. NRGene’s TraitMAGIC™ is a full turn-key solution for markers identification. It connects the dots between molecular markers, such as SNPs, and observed traits.
Quantitative trait loci mapping (“QTL mapping”) is an analysis method used to identify certain positions (loci) on a genome estimated to influence a certain trait, or phenotype. QTL mapping methods have come a long way in recent years. With TraitMAGIC™, we use the ultra-high-density genetic map to inform a highly accurate analysis that also provides each locus P-value—indicating the statistical estimate of the correlation.
TraitMAGIC - Genetic map and QTL analysis
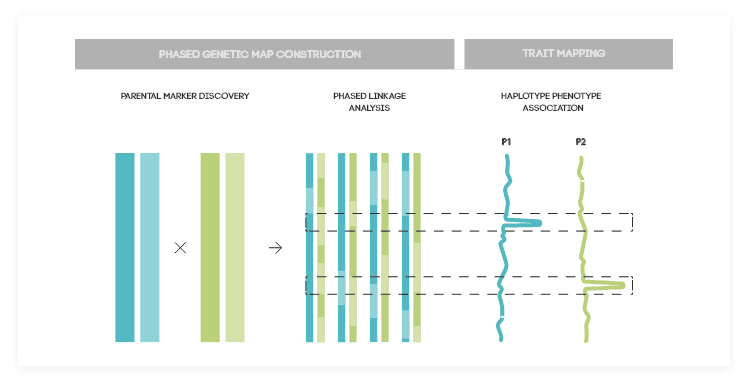
TraitMAGIC™ is NRGene’s revolutionary solution for genetic map construction. This technology brings together bi-parental population and QTL analysis to correlate genotype with phenotype. Allowing to see how DNA data leads to important traits.
TraitMAGIC™ uses a state-of-the-art technology based on haplotypes analysis. With this technology, and NRGene’s special algorithm, we can successfully analyze heterozygous and polyploid genomes with multiple recombination events.
The result is an ultra-high-density genetic map with millions of markers identifying recombination events. This genetic map can then be used to construct pseudo-chromosomes by the alignment of assembled genomic scaffolds to it. Combining the genetic map with additional phenotypic data, and using a highly accurate statistical analysis, can reveal quantitative trait loci (“QTL”) significantly correlated to a certain trait.
How it Works
AN EXTENSIVE YET SIMPLE TOOL FOR ANALYZING, STORING AND MINING UNLIMITED VOLUMES OF GENOMIC DATA
Outstanding Results
REAL CASE STUDY 1 (HOMOZYGOTE):
241 significant loci affecting 129 fruit quality traits were identified by converting over 58,000 markers to 3,663 haplotype bins in a melon RIL population*
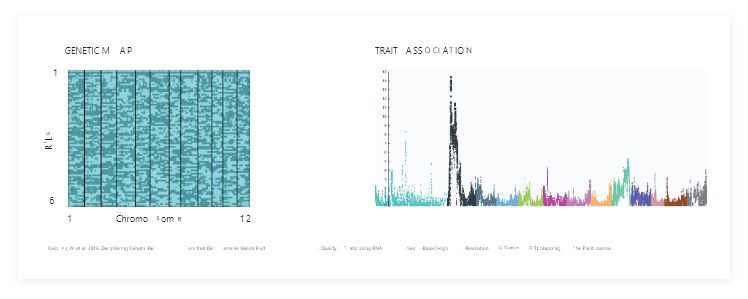
REAL CASE STUDY 1 (HOMOZYGOTE):
A phased haplotype approach (TraitMAGIC) was found superior to a consensus marker-based approach even when applied to <25% of a heterozygote F1 population
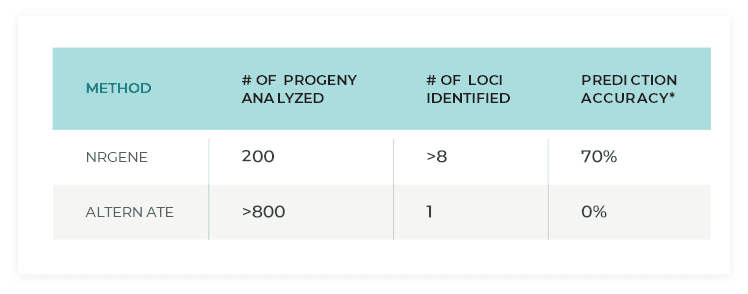
*percentage of progeny whose phenotype can be predicted with high accuracy based on the results.


